Covid Statistics
covid.RmdData Description -
This package provides the
covid_colsadataset which contains 35 variables for Covid stats in Colombia and South Africa from January 2020.The data comes from Guidotti, E., Ardia, D., (2020), “COVID-19 Data Hub”, Journal of Open Source Software 5(51):2376, doi: 10.21105/joss.02376, through the R package “COVID19” and filtered for the country Colombia and South Africa.
covid_colsa #> # A tibble: 542 x 35 #> # Groups: id [2] #> id date tests confirmed recovered deaths hosp vent icu #> <chr> <date> <int> <int> <int> <int> <int> <int> <int> #> 1 COL 2020-01-22 NA NA NA NA NA NA NA #> 2 COL 2020-01-23 NA NA NA NA NA NA NA #> 3 COL 2020-01-24 NA NA NA NA NA NA NA #> 4 COL 2020-01-25 NA NA NA NA NA NA NA #> 5 COL 2020-01-26 NA NA NA NA NA NA NA #> 6 COL 2020-01-27 NA NA NA NA NA NA NA #> 7 COL 2020-01-28 NA NA NA NA NA NA NA #> 8 COL 2020-01-29 NA NA NA NA NA NA NA #> 9 COL 2020-01-30 NA NA NA NA NA NA NA #> 10 COL 2020-01-31 NA NA NA NA NA NA NA #> # … with 532 more rows, and 26 more variables: population <int>, #> # school_closing <int>, workplace_closing <int>, cancel_events <int>, #> # gatherings_restrictions <int>, transport_closing <int>, #> # stay_home_restrictions <int>, internal_movement_restrictions <int>, #> # international_movement_restrictions <int>, information_campaigns <int>, #> # testing_policy <int>, contact_tracing <int>, stringency_index <dbl>, #> # iso_alpha_3 <chr>, iso_alpha_2 <chr>, iso_numeric <int>, currency <chr>, #> # administrative_area_level <int>, administrative_area_level_1 <chr>, #> # administrative_area_level_2 <lgl>, administrative_area_level_3 <lgl>, #> # latitude <dbl>, longitude <dbl>, key <lgl>, key_apple_mobility <chr>, #> # key_google_mobility <chr>
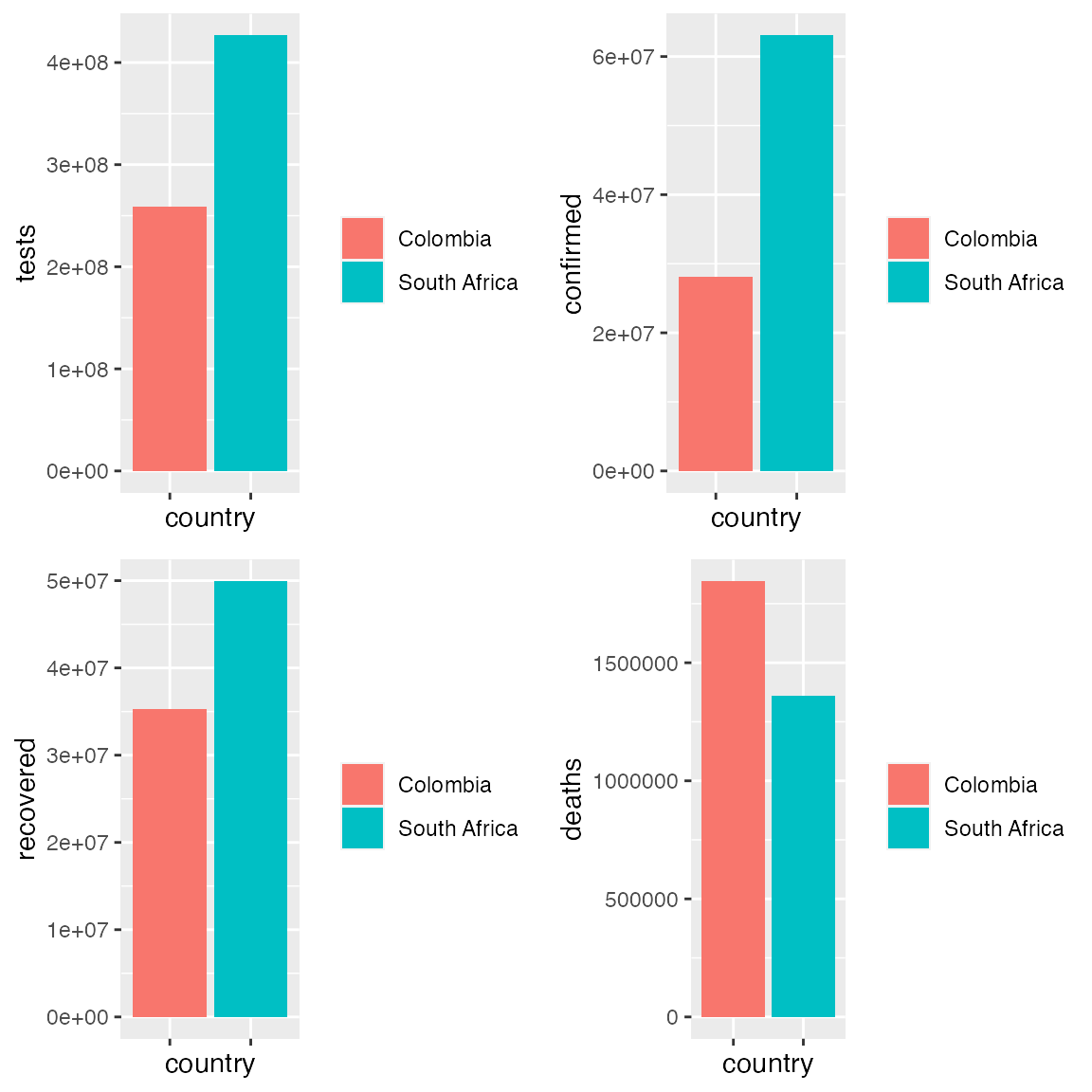
t <- ggplot(covid_colsa) + geom_bar(aes(x = key_apple_mobility, y = tests, fill = key_apple_mobility), stat = "identity") + theme(legend.title = element_blank(), axis.text.x = element_blank()) + xlab("country") c <- ggplot(covid_colsa) + geom_bar(aes(x = key_apple_mobility, y = confirmed, fill = key_apple_mobility), stat = "identity") + theme(legend.title = element_blank(), axis.text.x = element_blank()) + xlab("country") r <- ggplot(covid_colsa) + geom_bar(aes(x = key_apple_mobility, y = recovered, fill = key_apple_mobility), stat = "identity") + theme(legend.title = element_blank(), axis.text.x = element_blank()) + xlab("country") d <- ggplot(covid_colsa) + geom_bar(aes(x = key_apple_mobility, y = deaths, fill = key_apple_mobility), stat = "identity") + theme(legend.title = element_blank(), axis.text.x = element_blank()) + xlab("country") grid.arrange(t, c, r, d) #> Warning: Removed 89 rows containing missing values (position_stack). #> Warning: Removed 128 rows containing missing values (position_stack). #> Warning: Removed 91 rows containing missing values (position_stack). #> Warning: Removed 97 rows containing missing values (position_stack).